👋 to the 100+ new readers who joined since our last post. We’re back with part 2 in our series on LLMs for healthcare, a deep dive into use cases. Heads up, this one’s a longread.
2 trillion gigabytes of healthcare data are generated every year — the majority of which are unstructured.
Structured data are usable. You can store it in a database, load it into Excel, analyze it, generate charts, and share it easily.
Unstructured data — diagnostic reports, physicians’ notes, voice memos, and transcripts — are not so easy.
Historically, the best way to get information out of documents was to pay people to organize it (read: copy and paste it into Excel).
Old-school NLP systems could help somewhat. For example, you could train a model to tag words (“acetaminophen” → “Drug” and “cancer” → “Disease”). But medical terms have lots of synonyms (“acetaminophen” == “Tylenol” == “paracetamol”), so you’d still need to map tags to ontologies so the data can be searched or analyzed.
For these reasons, NLP had modest uptake in healthcare, until recently.
In the new age of language modeling, healthcare records and documentation are vastly more usable and useful. Let’s dig into how:
LLMs are expert data labelers and coders
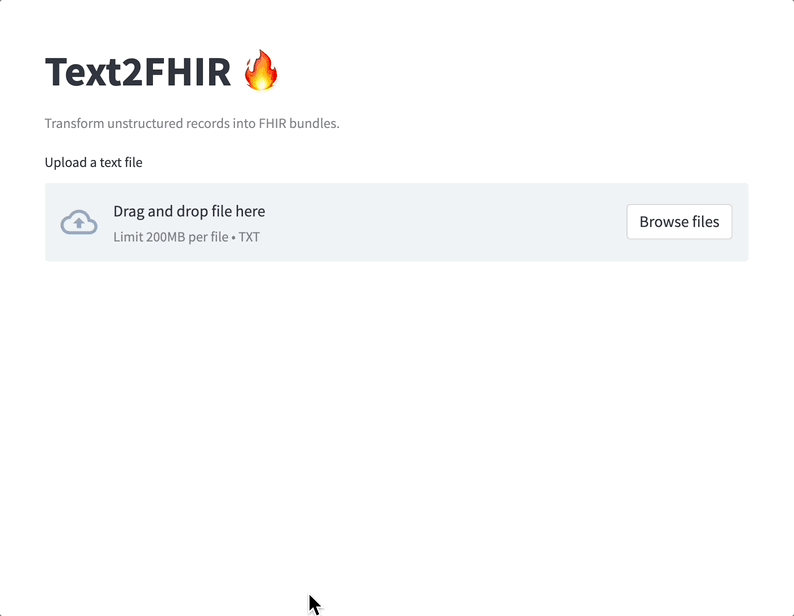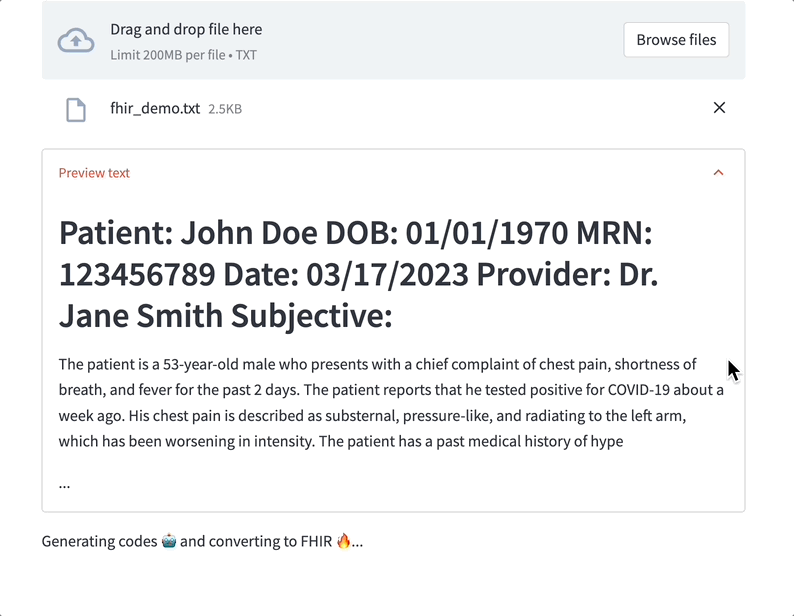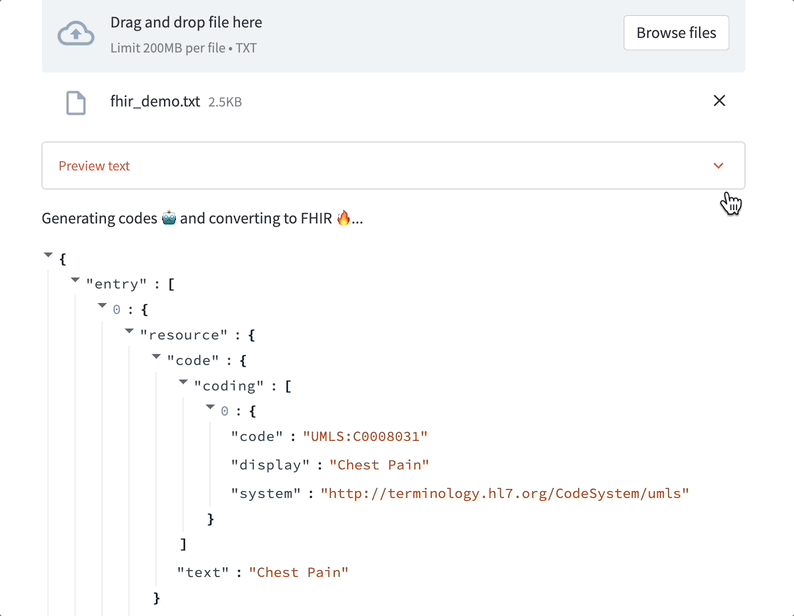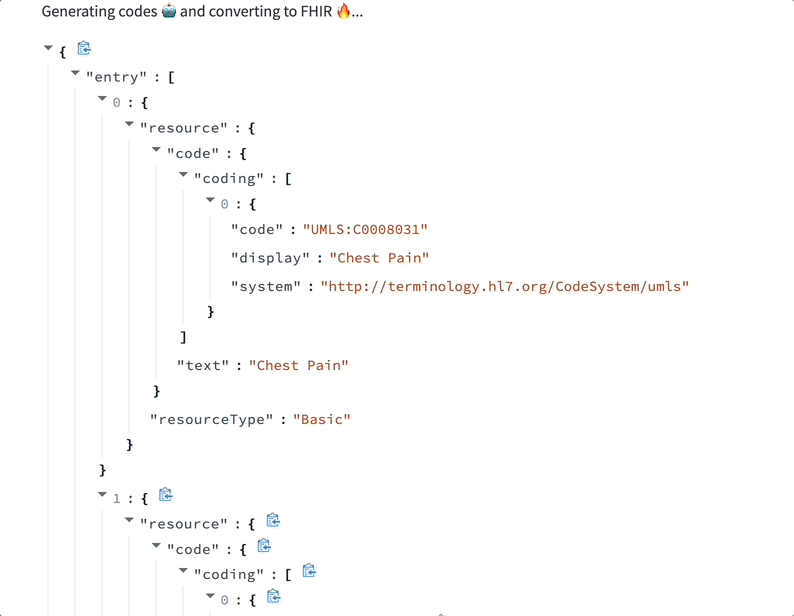
Models trained to label text using medical ontologies can transform documents into structured tables — faster than people can and with higher accuracy than old-school NLP or regex.
One of the first models we built was designed to find key terms in text and automatically map them to 20+ clinical ontologies in a few seconds. Given any medical document, it finds every medical condition, therapy, biomarker, and more. Our goal was to demonstrate that healthcare documentation could be richly annotated using LLMs better and faster than before.

Data recovered from unstructured text can improve our understanding of patients. Last year, researchers from MGH & the Broad Institute published a study where they used a BERT-based model trained on discharge summaries to recover 31% of missing patient data (height, weight, systolic, and diastolic blood pressure), resulting in a less biased dataset for analyzing patient outcomes.

Data labeling LLMs can assist with medical coding. Here’s a demo where a ScienceIO LLM turns the transcript of a physician's note into a list of medical codes:

Considerations:
Medical ontologies are often repetitive or redundant, especially in newer fields like precision medicine (UMLS has hundreds of entities related to a single gene). To build a reliable coding system, you need experts to curate what you want your model to train on and therefore produce.
Codes change frequently, particularly in billing, and thus the model must be re-trained or fine-tuned to keep pace.
If you want to use generative models for coding, know that they can “hallucinate”, or create fake codes that look real; Encoder models are more accurate and reliable but are harder to adapt to new codes than generative models.
LLMs are patient privacy tools
HIPAA violations carry significant fines and data breaches and attacks from bad actors can cost upwards of $10 million per incident. HIPAA anxiety has had a lot of unintended consequences, perhaps the worst of all being that it prevents a lot of teams from creating new solutions for patients out of fear of running afoul of HIPAA.
Trained properly, LLMs can find, redact, and flag protected healthcare information (PHI) or personally identifiable information (PII) much better than previous approaches (like regex).
We decided to develop our own healthcare-specific PHI LLMs after we heard from many customers that privacy tools built for other industries often removed vital clinical data in the process of redacting data. In one case, general-purpose privacy tools removed every abbreviated medical condition and biomarker in a customer’s data, rendering the “private” data useless.
Consider this note:
Will Roberts is a 65-year-old male, residing in Phoenix, AZ, 85004, with a history of urinary tract infections. He presented to the Pearson clinic today with symptoms of dysuria and frequency. A urine culture was performed by Dr. Jones and showed significant growth of E. coli. The patient was started on a course of oral antibiotics and will follow up with the clinic in one week for a repeat urine culture. If no improvement, the patient will be referred to St. Joseph Hospital.
An LLM trained to find and classify PHI/PII can output a table like this:
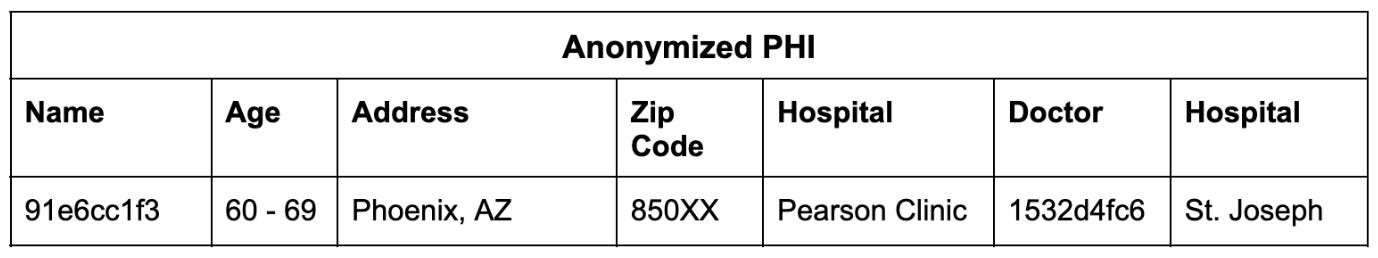
Once the PHI is detectable, you can anonymize the record so it can be used responsibly while maintaining the integrity of the clinical information:
In this way, we can link multiple records to an individual and then anonymize the totality of their records.
Considerations:
All algorithms have biases and LLMs are no exception. For example, they may fail to detect certain kinds of names (an early version of our PHI model misclassified every South Asian patient as a doctor, suggesting that the training data was curated by Desi parents). Therefore, bias testing is a critical part of our workflow.
We recommend not training PHI models with real patient information, especially generative models, because that can ironically pose a privacy risk to the patients in the training data. The jury is still out on how to responsibly use patient records to train LLMs (we have thoughts we’ll share in a future post!)
LLMs are mathematical matchmakers
"85% of all clinical trials fail to recruit enough patients," says Rosamund Round, Vice President of Patient Engagement at Parexel. "80% [of trials] are delayed due to recruitment problems and [high] dropout rates."
Recruiting and retaining a single patient for a clinical trial costs hundreds of dollars. And trials recruiting patients with rarer attributes, such as a very specific biomarker, will pay much more — sometimes over $1M per patient. The challenge is even greater for trials with medications that have specific inclusion criteria — a new precision oncology drug will have a much smaller targetable population than a general anti-inflammatory drug, and thus higher recruitment costs per patient.
LLMs can help trial sponsors and sites expand the potential pool of recruitable patients by matching trial criteria to patient attributes in the EMR.
There are a couple of ways to do this:
LLMs trained using clinical information can represent both patient profiles and clinical trial criteria as vectors. Using these vectors, one can generate a list of candidate patients for a trial, or candidate trials for a patient very quickly.
Or, LLMs can find key terms in patient records to determine if they are a match (biomarkers, diagnostic criteria, vitals, test results). One benefit to this approach is that the “match” is more interpretable for a patient — you can search for patients based on specific observations, which a vector-based method cannot easily allow.

Last year, we used an LLM to analyze thousands of de-identified patient records on the Mayo Clinic Platform. Within 24 hours we found 55 patients who qualified for selective triple-negative breast cancer trials. None of these patients could be found by searching by ICD-10, CPT, or LOINC codes related to cancer.
Making trials visible to the patient or the care team can help solve for the top of the trial recruitment funnel, but only a small fraction of patients will enroll.
To increase the probability of enrollment, generative LLMs can help with patient engagement. LLMs can provide clear explanations of the clinical trial’s goals, the candidate drug, how it works, what the trial entails, and other resources to engage patients and care teams. They can describe why the patient is a candidate and draft a note for the patient, explaining the personal opportunity and the impact they can make by enrolling.
A recent study in JAMA Internal Medicine had licensed healthcare professionals grade responses to patients’ questions, pitting ChatGPT against real physicians. They found that ChatGPT’s explanations were longer, higher quality, and more empathetic. This makes sense, ChatGPT is partially trained using peoples’ feedback on the quality of its outputs, so part of its goal is to appeal to users’ preferences. That these preferences on general tasks translated to a clinical context is only mildly surprising.
Still, I don’t recommend replacing doctors with chatbots. Instead, patient-centric solutions can have the best of both worlds — an LLM can generate possible responses, and care teams can select those that are accurate and empathetic. Some cognitive burden is transferred to the model, and communication is still between the patient and their carer.
Considerations: A matching algorithm may have biases towards or against biases certain patient populations, indications, or even therapeutic modalities. These biases will impact the diversity and fairness of trial recruitment. We recommend testing any algorithm with data (real or synthetic) representing different patient populations to identify biases before real-world application, and ongoing.
LLMs are tireless automation machines
Administrative costs are estimated to be ~20% of national healthcare spend, or ~4% of the US GDP.
At a panel discussion in 2018, Donald Rucker, the former national coordinator for US Health IT, described a finding from a study he performed at Ohio State. “Of the 130,000 phone calls a day made by that 800-bed health institution, roughly half were 60 seconds or less, exchanging one fact.” Rucker envisioned EMRs as looking more like enterprise software where information exchange is less manual and more seamless.

LLMs are excellent at creating templated documentation. Reliable generative models can allow administrators to quickly draft information to be transferred, or even populate structured data fields from transcripts or voice memos.
As LLMs get increasingly plugged into other software and we get more tools that let them tap into data sources and APIs, they will become even more useful for automation. We’re rapidly heading towards an API-first health ecosystem, and that’s awesome for all the new data we’ll make, but what about the data that’s piled up? Translating between systems is tedious work, and no person wants to do it. LLMs acting as file-to-API and API-to-API translators can smooth out some of the rough edges.
Considerations:
Because healthcare information is sensitive and needs to be highly-accurate, human supervision and reinforcement are necessary to achieve maximum quality control.
I don’t see a future where massive GPT-X models are used universally across all use cases. Instead, smaller models trained specifically for complex tasks which are easier to integrate into a private setting and easier to fine-tune will prevail. For example, automated coding is possible with LLMs, but you want to use a model specifically trained on medical ontologies since, as we mentioned, general-purpose models often hallucinate codes.
LLMs are memory machines
On average, a physician in the US consults with 2,000 patients each year.
Patients can have hundreds of pages of unstructured data and electronic health records, much of which can be repetitive. Many do not have the time and resources to absorb the full context of each patient’s history, let alone stay up to date on clinical trials and new research studies.
Fortunately, LLMs are highly effective data compression machines.
Transformers are designed to encode lots of information compactly and efficiently, and they must learn to decode that encoding to produce coherent and contextually appropriate responses. Even smaller LMs are able to learn from >1TB of text and produce weights in the GB regime.
This means LLMs can act as user-friendly knowledge bases. They provide information from the text they’ve been trained on and can even do so in different styles or languages.

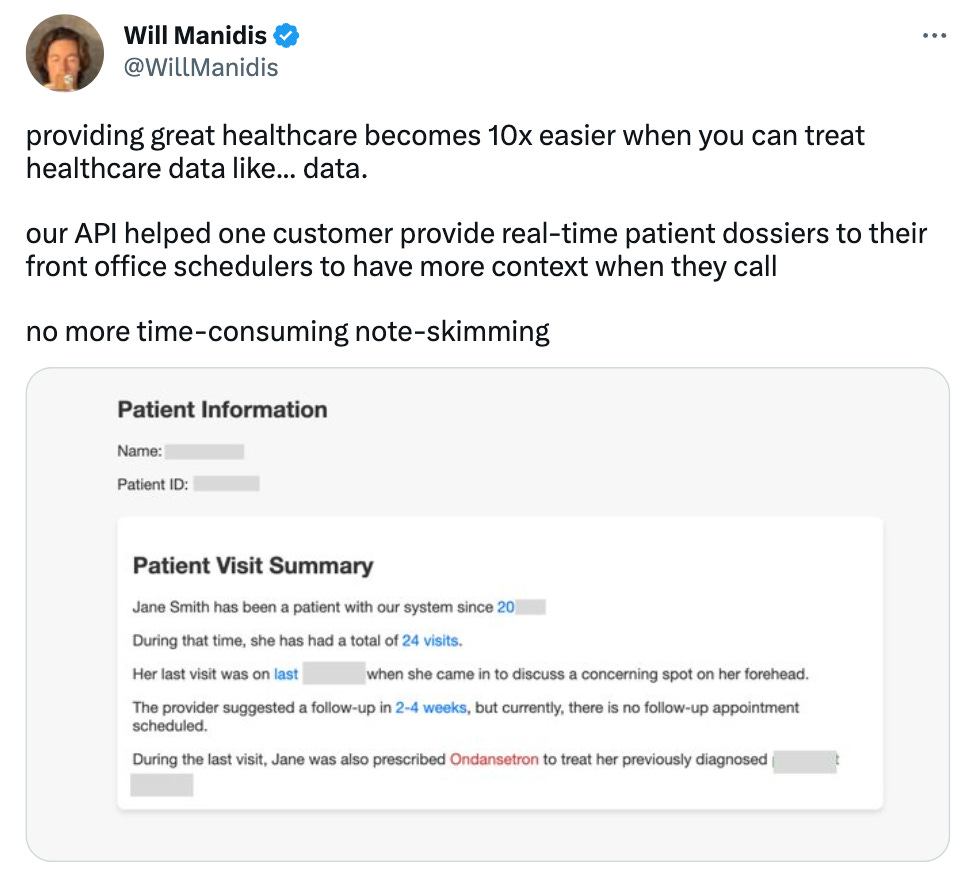
When provided with context, LLMs can also extract relevant information from it. This is particularly beneficial in the context of patient information retrieval. LLMs with the ability to pull data from records can assist in clinical workflows by summarizing a patient’s history, providing key data, and recommending potential tests or treatments based on its knowledge of the patient, treatment protocols, and research studies.
Here’s an example where LLMs were able to provide a highlighted patient summary. A generative model produced the summary, and our encoder-based LLM detected all the key terms:
There are a lot of considerations to be made when relying on LLMs for information:
As compression algorithms go, LLMs are lossy — they will not perfectly preserve all the original details of the text they were trained on and can produce inaccurate or imprecise answers. After all, many LLMs are only trained to produce coherent text, not necessarily accurate text. Therefore, we do not recommend taking a model’s output at face value without confirming a reliable source.
Here’s a personal example: when I’ve used GPT-4 for literature search of arXiv, >99% of the papers it gives me are real but only ~40% of the source URLs are correct (~60% point to real but unrelated papers).
Hallucination can be mitigated in many ways, including giving LLMs access to external databases, writing code to validate links, and/or rewarding them when they provide accurate sources. Keep an eye on how search engines are integrating LLMs and knowledge graphs to deliver more accurate results.
LLMs are people simulators
LLMs can build a “representation” of individual people mentioned in its training data. Certain words are more likely to be associated with a person, and this statistical probability is encoded into the model.
This is why LLMs can provide relevant information on a person, like describing their biographical history or notable attributes — the words it generates are more likely to appear in the source text.
LLMs can also draw upon the shared attributes of individuals when generating text:

Thanks to this property, LLMs can serve as a statistical model informed from each patient’s experience. When trained on patient records, they will form a representation of each patient, doctor, and other individual in the records.
Learning from each patient's experience is challenging. Physicians take years to build an intuition for how to diagnose and treat patients. This intuition is refined with each patient encounter and as medicine advances. LLMs approximate that process in a fraction of the time, and may develop emergent capabilities which might be useful for patient care.
What would you do if you could draw on the history of a million patients in real-time?
For a long time, I’ve been fascinated by the idea of a patient atlas — an algorithm that, when given a limited amount of info about a patient, could produce a differential diagnosis, assign probabilities to each diagnosis, suggest interventions, tests, or treatments, and refine its working model of them as it learns more.
Given the rapid pace of artificial intelligence, I don’t believe these capabilities are far out.
Considerations:
LLMs trained on real patient data will remember real patient details, which poses a serious privacy risk to those patients. This problem is much worse when you train a neural network on tabular data (even a modest GAN can fully memorize most clinical datasets in as little as 2 epochs). Anonymized datasets covering a wide swath of the patient population will be crucial.
It’s important to remember LLMs are only learning from signals that are documented, whereas physicians often pick up on nuanced signals that no EHR captures.
In the near term, I believe LLMs will be most useful in reducing cognitive burden for care teams, patients, operators, and developers. LLMs save time by performing long and tedious that require no special expertise, and they can save energy by supporting quick tasks that require mental effort.
Longer term, I genuinely believe that we can reshape the way healthcare works, and AI-driven products purpose-built for healthcare will play a big part. In our post on the last 15 years of healthcare, we reflected that healthcare is capable of incredible transformation given the right conditions. LLMs are a powerful tool we can use to improve healthcare if we think about how to do the right jobs, not just do the same ones faster or cheaper.
That’s the change we build for.
Thanks for reading,
Gaurav 🌊
Huge thanks to Dennis Ryu, Alda Cami, and Will Manidis for their feedback. 🙏🏽







One way we've seen the PHI problem avoided is generating synthetic equivalents of the data you're trying to train on. So when you have an output table like you showed with age/address/etc., you can start with those variables and then use LLMs to generate the transcripts/reports that would reflect that patient.
This doesn't help you gain new information from data, but it does help you build a model better able to do complex medical NER style tasks like the ones you were mentioning!